Retrosynthesis tools review
The ability to identify manufacturing pathways for given products is of the highest importance to researchers, private organizations, and societies as well. In an era of endless theoretical predictions of new medicine, new active and functional chemicals, and new materials, the question that comes up is ONE:
‘HOW do we synthesize the chemical that we predicted as the solution to a given problem?’
The question has more applications, even for known compounds with known synthetic pathways. Different questions can be the case:
‘how can I synthesize a chemical with a known but patented synthetic approach?’
‘how can I synthesize a chemical with an alternative process which is of lower cost?’
‘how can I synthesize a known chemical with an alternative process that uses materials that are available locally?’
The answer to the above question is termed retrosynthesis. Retrosynthesis allows for the suggestion of alternative synthetic pathways for given targets (compounds of interest) based on mainly two (three including the hybrid) approaches:
a- The rule based approach
b- The data- driven approach
c- Hybrid approach
In the rule based approach, chemical rules and reaction classifications are imported/ programmed into the tool, and thus the retrosynthetic suggestions are based on knowledge imported by the creators of the tool. On the contrary, the data- driven approach allows the tool to ‘learn’ from variant data sets, and thus being able to either focus on specific cases or always learn ‘new chemistry’. There are advantages and disadvantages to each of the methods, and the hybrid approaches attempt to combine the best features of both.
During our involvement in the retrosynthesis project with a very known innovative pharmaceutical, we had the chance to work and evaluate a series of state of the art retrosynthesis tools, that included both commercial and free ones as listed below:
n RetroPath 2.0- a reaction rules based tool, that is freely available and incorporated into Galaxy as well. Rules have been imported with the use of Chemaxon Marvin Sketch manually. Reactants and reactions have been acquired by KEGG, Metacyc, Rhea, and Reactome.
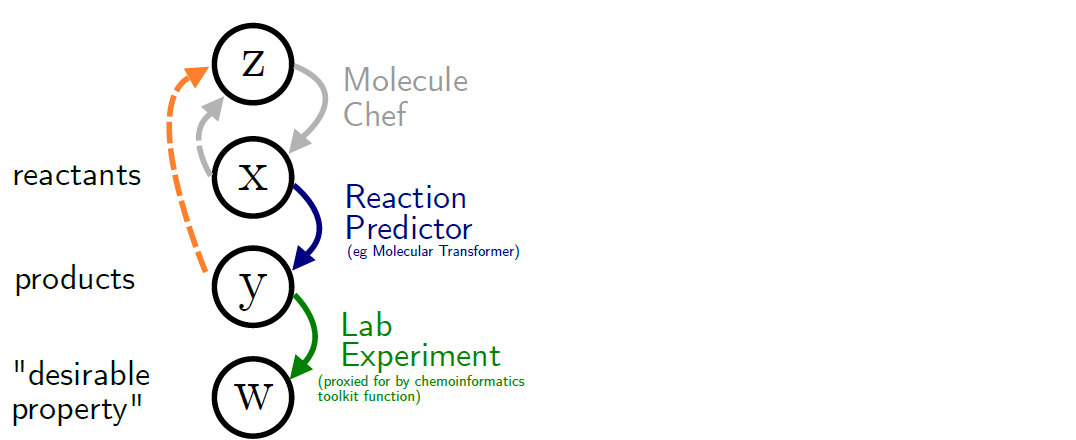
n A molecular generator that can be used as a potential retrosynthesis tool is MOLECULE CHEF. The MOLECULE CHEF is proven to be quite efficient in generating structures. MOLECULE CHEF functionality can be reversed when coupled with a regressor to perform retrosynthesis
n Retroformer is a transformer based architecture that introduces the ‘local attention head’. Its aim is to boost the reaction reasoning ability of deep generative models in retrosynthesis. The top-1 accuracy reaches high scores of 64 and 53.2%, for reaction class known and unknown settings, respectively. Retroformer also improved the top-10 molecule and reaction validity by 23.6 and 22.0%, respectively compared to vanilla retrosynthesis transformer.
n One of the most suitable and successful retrosynthesis tools is SYNTHIA (previously known as Chematica). SYNTHIA includes 100,000 reaction rules that are recursively applied to the target compounds. In every reaction rule, ‘dynamic’ information is included about reaction conditions, conflicts among functional groups, etc.
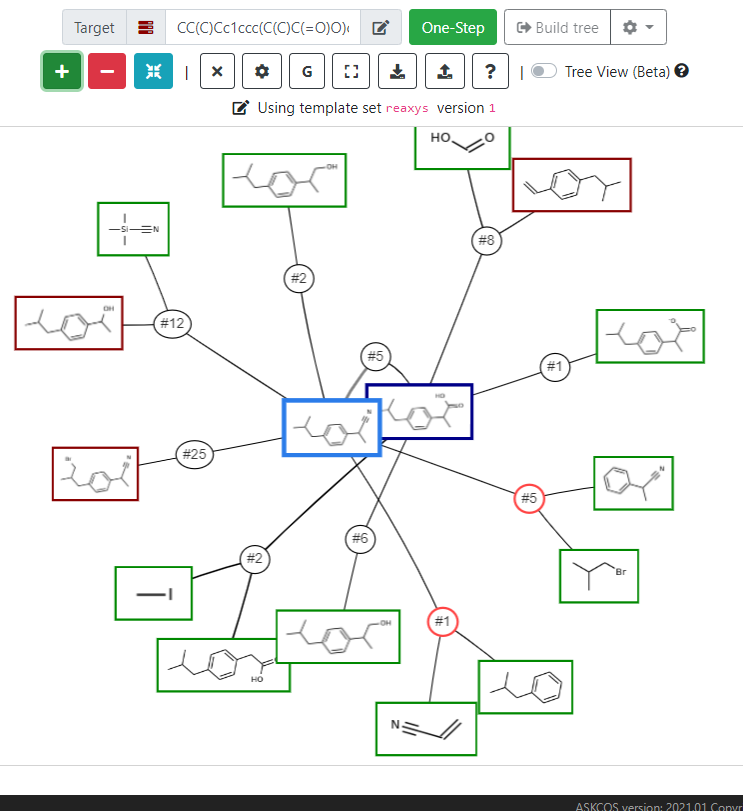
n ASKCOS suite is another open-source suite that is capable of reaction planning and one step retrosynthesis as well. ASKCOS can extract reaction templates from various databases (US Patent, Reaxys, etc.), and thus had achieved a library of 163K+ transformations. Different modules in the software attempt reaction prediction, likeness of final product, etc. The retrosynthesis module is accessible at Interactive Path Planning (mit.edu).
n AiZynthFinder is an open source retrosynthesis tools, written in Python 3 that is available at GitHub under the general MIT license. The overall programming is based on an object oriented programming approach. It offers complete searches in under 1min, and solutions in around 10 seconds. It is based on Monte Carlo tree searches, where each node corresponds to molecules/ fragments that can or cannot be further broken down into smaller ones.
n DDRAM/ ChemPrint is another combination of tools that can be used for retrosynthesis of drug (and not only) compounds of interest. It is based on mining millions of reactions (mainly from Eli Lilly library, but not only from there) and proceed to the development of reaction classes.
n ICSynth, now at DeepMatter, is another (proprietary) algorithm that allows for retrosynthesis of drug compounds, resulting to known and novel pathways. The current information available is not as accessible as it was in the previous years, and that could be due to current critical developments being made
n Galaxy- SynBioCAD is an online tool that allows for molecule retrosynthesis, in a very comprehensive way to the user. Registration is free, and a number of tools is available on the platform. The user can download or upload structure information, convert formats, assign jobs to the retrosynthesis module, get statistics on the jobs run, get analytics on the proposed pathways, etc.
n SensiPath (at SensiPath (micalis.fr) ) is another tool with a very easy GUI, and a known functionality that is mainly used for synthetic pathways in the metabolic space. The target compounds are represented in InChls and then converted in molecular signature for subsequent matching in the databases. Solution times are usually just a few seconds
n IBM RXN is easily one of the top 3 retrosynthesis tools examined. Like AKSCOS, it is free for registered users, so it was evaluated for a number of potential targets of interest. Inputs can be drawings or SMILES, with the drawing tool being easy and fast to use, even if you are not an experienced user. The time for solutions and complete answers is comparable to ASKCOS
n Reaxys makes use of some of the largest datasets available, utilizing 15M of reactions with 400,000 reaction rules. In addition to that, the software allows for further training on additional datasets, if they become available. Reaxys speed is also comparable to IBM RXN. Input is SMILES, InChiKey, Chemical name, CAS, and via a 2D drawing tool, similar to competition (ASKCOS, IBX RXN). InChiKey option allows for better treatment of regio – selective cases and stereo chemistry. Drawing capabilities come from Marvin JS/ ChemAxon.
n CAS SciFinder is the American Chemical Society’s competition to Reaxys. In the same manner, they utilize their vast information database to create 150,000 reaction rules in house, and thus being able to propose novel synthesis routes for existing and non- existing molecules. Their capabilities are very similar to Reaxys’.
n Chemical.AI (ChemAirs) is the next proprietary algorithm for retrosynthesis. In this case, besides the similarities to Reaxys and SciFinder (Inputs with 2D drawing tool, SMILES, CAS; 100K+ reaction rules; node inspection; filters of synthetic routes; presentation of results; etc.), a major breakthrough is found: the software is one step ahead of Reaxys (and SciFinder) in terms of dynamically altering the suggested synthetic routes (see note above for the next release of Reaxys).
n Spaya also accepts SMILES and 2D drawings as target inputs, lacking some types when compared to the competition. Spaya offers the ability to specify desired intermediates for the retrosynthetic steps, a feature that can also be of value to many users. In the same manner, the user can ask for specific reaction(s) to be included in the suggested retrosynthetic step
Our involvement has led to successful adaptation of retrosynthesis tools, and the initiation of an in house retrosynthesis tool.
